Using Wobble Rules, What Is the Minimum Number of Trna Needed to Read the Genetic Code
Abstract
Posttranscriptional modifications at the wobble position of transfer RNAs play a substantial role in deciphering the degenerate genetic code on the ribosome. The number and diversity of modifications suggest different mechanisms of activeness during messenger RNA decoding, of which merely a few were described so far. Here, on the basis of several 70S ribosome complex 10-ray structures, we demonstrate how Escherichia coli tRNALys UUU with hypermodified 5-methylaminomethyl-2-thiouridine (mnmvs2U) at the wobble position discriminates between cognate codons AAA and AAG, and near-cognate stop codon UAA or isoleucine codon AUA, with which information technology forms pyrimidine–pyrimidine mismatches. We show that mnmfives2U forms an unusual pair with guanosine at the wobble position that expands full general cognition on the degeneracy of the genetic code and specifies a powerful part of tRNA modifications in translation. Our models consolidate the translational fidelity mechanism proposed previously where the steric complementarity and shape acceptance dominate the decoding mechanism.
Introduction
More than a hundred of posttranscriptional RNA modifications identified todayane were shown to play diverse and indispensable roles in cistron regulation in all domains of life2. Modifications of RNA are carried out by complex cellular pathways, which involve countless protein enzymes and catalytic RNA–protein complexes, which primarily target tRNAs and, to a lesser extent, ribosomal RNA and mRNAs1. The observed trends advise that many modification motifs and their sequence locations are conserved throughout Bacteria, Archaea and Eukarya; however, some kingdom-specific differences are documented likewise.
Among all modifications found, those of tRNA are the most abundant and studied classes of modificationsane,3,4,5. Xc-iii tRNA modifications described today represent an astonishing library of chemically diverse structuresv each of which influences in a unique way three-dimensional integrity of the tRNA and specifies its physicochemical properties6,7. The most elaborate tRNA modifications are located in the anticodon loop, which comprises the anticodon triplet necessary for pairing with mRNA codons. The anticodon loops of almost all tRNAs contain several modified nucleotides. Amongst these, the about important are nucleotides in the positions 34 and 37. The nucleotide in position 34 (so-chosen 'wobble' position) pairs with the third mRNA codon base in the aminoacyl-tRNA binding site (A-site) during decoding4,8. The nucleotide in the position 37 is adjacent to the iii′-side of the anticodon.
From the moment of their discoveries, modifications in tRNA anticodon loops were demonstrated to be crucial for proper mRNA decoding and fine-tuning of the processnine. In particular, modifications of tRNA position 34 were implied to increase tRNA capacities to decode multiple mRNA codons differing past the tertiary nucleoside (synonymous codons), hence explaining how the degenerate genetic lawmaking is translated4,10,11. It was shown also that anticodon modifications enhance recognition by respective aminoacyl-tRNA synthetases12,xiii and serve as a preventing measure of frame shifting during translocation14,xv. Very recently, the tRNA modifications were too credited a role in connecting translation, metabolism and stress response in bacteria and eukaryotes2,16. For example, in humans the deregulation of RNA modification pathways was shown to be linked to the blazon ii diabetes and several mitochondrial diseasessixteen.
Except for methionine and tryptophan, all amino acids are encoded past more one codon, because the genetic code is redundant17,xviii with 61 codons encoding 20 amino acids. In dissimilarity to eukaryotes, where synonymous codons AAA and AAG are read by three isoacceptor lysine tRNALys, half of all bacteria have but one isoacceptor tRNALys UUU that decodes these 2 codons into amphipathic amino acrid lysineix,nineteen,20. To discriminate against pyrimidine-catastrophe codons AAC and AAU encoding asparagine, the E. coli tRNALys UUU contains one of the nigh complex modifications 5-methylaminomethyl-two-thiouridine (Due south, mnm5s2U) at the wobble anticodon position 34 (Fig. 1a). The mnmvsouth2 modification is a result of a sophisticated pathway that includes many enzymes responsible for thiolation and attachment of a methylaminomethyl grouping21. Another prominent characteristic of E. coli tRNALys SUU is N6-threonylcarbamoyladenosine (thalf-dozenA) at the 37th position of its anticodon loop (Fig. 1a). The t6A modification is i of the almost ubiquitous and conserved, and is known to be critical for recognition of codons starting with adenosine. This is one of the rare modifications universally conserved throughout different kingdoms of lifei. In add-on to S34 and t6A37, E. coli tRNALys SUU anticodon loop bears the third modification, pseudouridine at position 39 (Fig. 1a).
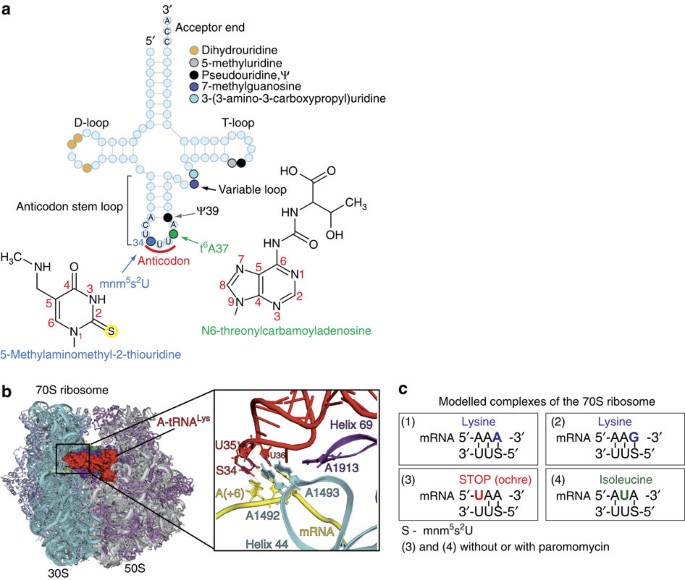
(a) Secondary structure of Due east. coli tRNALys SUU. Major domains and modifications of tRNA are indicated; chemical formulas of hypermodified nucleobases at positions 34 and 37 are given together with corresponding abbreviations. (b) Side view of the 70S ribosome complex with three tRNAs bound at the A- (ruby-red), P- (bluish) and get out (light-green) bounden sites. Helix 69 of the large subunit is in magenta. The frame designates the decoding heart with the leap anticodon-stalk loop of tRNALys SUU and the close-upwards view on the codon–anticodon duplex and major nucleotides of the decoding eye (G530 from 16S rRNA is not shown) including A1913 from 23S rRNA. (c) Schemes of codon–anticodon duplexes in the decoding centre of the 70S ribosome complexes modelled in the study. The complexes are numbered in accordance with description in the main text.
Nuclear magnetic resonance studies of unmodified, partially and fully modified anticodon stem loops (ASLs) of Eastward.coli tRNALys SUU demonstrated that mnmfivestwoU and thalf-dozenA modifications remodel an otherwise dynamic loop to canonical open U-plow structure to perfectly suit in the ribosomal decoding centrehalf dozen,22. First, X-ray studies of the partial decoding system23, where crystals of the isolated small ribosomal 30S subunit were soaked with synthetic ASL of E.coli tRNALys UUU and hexaribonucleotides as mRNA analogues, shed some light on possible roles of modifications in decoding24,25. It was suggested that tviA37 enhances the codon–anticodon stability via cantankerous-strand stacking interaction with the first codon nucleotide, whereas partially modified mnm5U34 lacking 2-thio group was implicated in an alternating mnmfiveU34·M base-pairing interactions via a bifurcated hydrogen bond25. A similar model of the bacterial 30S subunit with ASL of human tRNA3Lys UUU, which has identical anticodon loop sequence with Due east. coli tRNALys SUU just carries msiit6A37 (2-methylthio-N6-threonylcarbamoyladenosine) and mcm5s2U34 (5-methoxycarbonylmethyl-2-thiouridine) modifications, revealed Watson–Crick-like geometry of mcmfives2U34·Thou base-pairing interactions at the wobble codon–anticodon position26. Both of these works and numerous other genetic, biophysical and biochemical findings indicated that each group in a modified nucleotide improves thermodynamic backdrop of tRNA and serves to augment specific codon–anticodon interactions during decoding.
Contempo crystallographic studies of the 30S ribosomal subunit or complete 70S ribosome complexes with different ASLs23,24,25,26,27 or tRNAs28,29,30,31,32,33 and mRNAs describe 3 major classes of preferred geometry at the wobble position of a codon–anticodon minihelix. The get-go class consists of canonical Watson–Crick purine–pyrimidine A·U (or U·A) and C·Yard (or Chiliad·C) pairs29,30,33, and also includes Watson–Crick-like pairs. Information technology was suggested that the latter closely resemble approved geometry via stabilization of an enol tautomer past the wobble modifications (for example, in a higher place described mcm5siiU34·Thousand)26,27. The second class consists of standard wobble pair G34·U originally predicted past Crick8. In this pair, the pyrimidine is displaced towards the major groove of the codon–anticodon minihelix; however, the distance between the ribose C1′ atoms remains very close to 10.v Å, the boilerplate value for a standard Watson–Crick pair28,29. The third grade includes I·G or I·A pairs, where I (inosine) is present at position 34 of some tRNAsone,34. These purine–purine pairs are unusually wide with a C1′–C1′ distance of 12.3 Å (refs 24, 32).
In the current report, we describe six Ten-ray structures of physiologically relevant complexes of the complete 70S ribosome primed with long mRNAs that comprise full-length native E. coli tRNAfMet in the peptidyl-tRNA-bounden site (P-site) and tRNALys SUU leap to cognate or almost-cognate codons in the A-site. We identified an unprecedented base-pairing interaction at the wobble position of the codon–anticodon duplex in the decoding eye that broadens the nowadays family of 'wobble geometries'. This base pair, which involves a hypermodified S34 of Due east. coli tRNALys SUU and codon guanosine, represents a 'wobble' (G34·U) with the U moved towards the minor instead of the major groove that is much less isosteric to its flipped form than usual wobble Yard·U pair.
The ribosome structures we are describing in this piece of work deepen the understanding of the tRNA discrimination mechanism on the ribosome. We demonstrate how tRNALys SUU discriminates between cognate codons AAA and AAG, and the near-cognate stop codon UAA (ochre codon) or the isoleucine codon AUA, with which information technology forms pyrimidine–pyrimidine U·U mismatches. Together with our earlier structures of the 70S ribosome with various mismatches in the codon–anticodon duplex29,thirty, the present models expand our library of various states of the 70S decoding centre. The present evidence further strengthens our proffer that the steric complementarity is predominant over the number of hydrogen bonds35 betwixt the decoding center and the codon–anticodon duplex, and hence plays the crucial discriminatory part during decoding.
Results
The modifications of lysine tRNA in the 70S decoding centre
In this work equally in our previous studies28,29,xxx, nosotros employed the total Thermus thermophilus 70S ribosome co-crystallized with long synthetic mRNAs and natural tRNAs. These complexes model cognate or well-nigh-cognate states of the decoding centre at the proofreading stride of the tRNA pick process (Fig. 1b). Nosotros adamant six X-ray structures of the 70S ribosome programmed by 30-nucleotide-long mRNAs with AUG codon and E. coli tRNAfMet in the P-site and the A-site occupied by tRNALys SUU jump to its cognate codon AAA or AAG, or virtually-cognate stop codon UAA and isoleucine codon AUA (Fig. 1c and Tabular array 1).
The electron density maps of the 2 cognate complexes (Fig. 1c, complexes 1 and 2) possess sufficient level of particular to discern structural features of the tRNA anticodon loops that can be attributed to the influence of modifications mnmvsouth2U and t6A (Fig. 2). The amino grouping of the modified nucleotide S34 forms a hydrogen bond with 2′-OH of nucleotide U33 (Fig. 2a), thus altering the U-turn structure and stabilizing it, as was shown before for isolated ASLs6. The thio group of the aforementioned nucleotide is known to stabilize 3′-endo conformation of the ribose favourable for base-pairing interactions36, influencing the codon–anticodon helix stability. We observed a characteristic, characteristic of 2-thiouridine37 as well, namely the S2(S34)-N1(U35) 'stacking' interaction with the subsequent nucleotide U35 (Fig. 2a). This interaction affects the relative positioning of the S34 and U35 nucleotides, and hence the shape and stability of the codon–anticodon duplex.
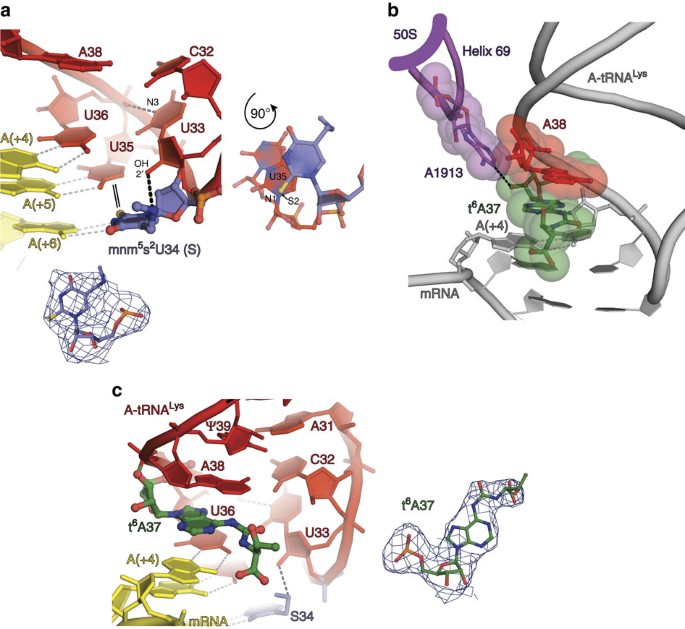
(a) The S34 interactions: stabilization of the U-plow structure via anchoring of the mnmfive group and stacking of the thio group with the N1 atom of U35 shown in an culling top-view orientation on the right. Nucleotide 37 is omitted for clarity of representation. (b) The invariant A1913 in Helix 69 of 23S rRNA of the big ribosomal subunit (thick magenta line) constrains position of the t6A37 ribose by conserved hydrogen bail interactions; van der Waals surfaces show that A1913 besides defines conformation of the anticodon loop from the three′-side of t6A37 at position 38. (c) Cross-strand stacking of t6A37 with the first nucleotide of the A-site jump codon (position (+four)). In a and c, 2F o−F c electron density maps corresponding to S34 and thalf-dozenA37 are contoured at one.0σ.
An influence of the large 50S ribosomal subunit on the tRNA constraints during decoding remained underestimated for a long fourth dimension, because the first models of decoding were based on the structures of the isolated pocket-size ribosomal subunit23,35. On the 70S ribosome, the conserved helix 69 of the 50S subunit, which is pivotal for many functions of the ribosome, directly contacts the carbohydrate moiety of the tRNA nucleotide at position 37 (refs 31, 38 and Fig. 2b). Nigh probably, this contact is important for proper positioning and conformational stabilization of the anticodon loop. In addition, t6A37 forms cross-strand stacking with the first nucleotide of the mRNA codon in the A-site (Fig. 2c). Similar stacking interactions were described in the early models of the 30S subunit whose crystals were soaked with the tRNALys UUU ASL carrying thalf dozenA37 and mnmfiveU34 modifications25. However, the comparison of our nowadays structure with this model revealed a considerable shift of 1 Å in the position of the t6A37 nucleotide pointing to a specific role of the big 50S subunit in restraining the anticodon loop of the A-site bound tRNA (Supplementary Fig. one). The position of tsixA37 over the A·U pair itself (Fig. 2c), as observed in our structures, corresponds well with the main function of this modification in stabilizing the weak A·U base-pairing interactions and preventing mRNA slippage during translocation equally well.
Hypermodified uridine forms a unique base pair
The capacity of E. coli tRNALys SUU to read both codons AAA and AAG catastrophe with purines implies that the hypermodified uridine S34 at the first anticodon position is involved in a dual mode of base-pairing interactions with adenosine and guanosine. In general, in bacteria the AAA codon is used approximately iii times more than often than the AAG codon39. However, information technology was estimated that when the side by side codon later the ane encoding lysine starts with cytidine the AAG codon becomes preferred40.
Our first structure with E. coli tRNALys SUU bound to its cognate AAA codon showed that hypermodified uridine S34 formed a distorted Watson–Crick base pair with the opposing adenosine with the standard C1′–C1′ distance of 10.6 Å (Fig. 3a). The slight positional difference of the uracil ring from its standard position in the Watson–Crick A·U pair, caused past the interactions of the modifications with neighbouring nucleotides U33 and U35 (Fig. 2a), resulted in weakening of interaction between the codon adenosine and S34. On the other paw, the very same interactions tend to strengthen the codon–anticodon duplex equally a whole past adjustment of the shape of tRNA U-turn6 (Fig. 2a).
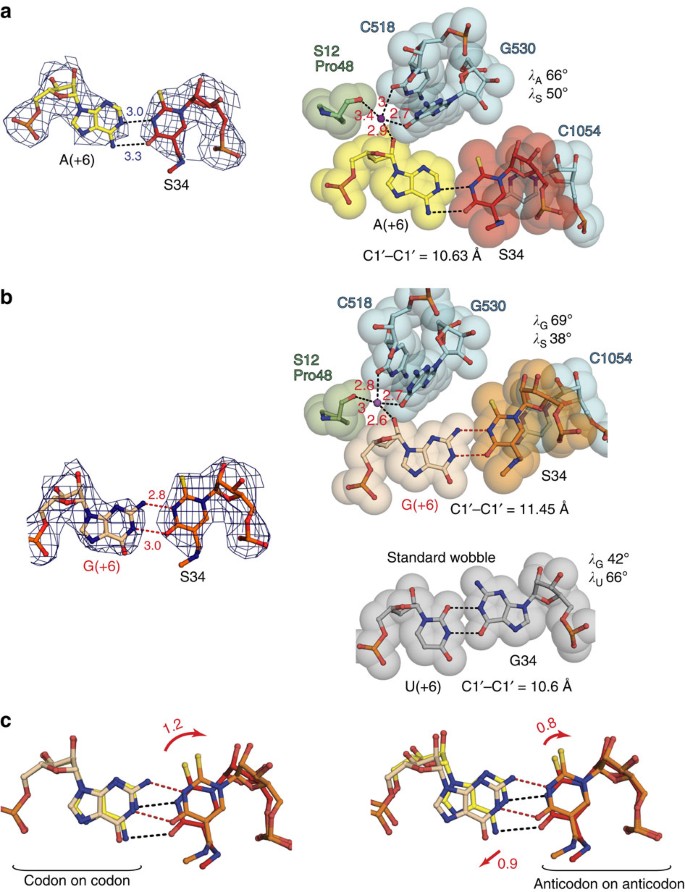
(a) The S34·A(+half dozen) pair. Left, hydrogen bonds are indicated (Å); right, van der Waals surfaces with the corresponding C1′–C1′ distance and glycosidic angles (λ). (b) The S34·G(+half dozen) pair; indicated parameters every bit in a; possible hydrogen bonds are marked by red dashes. In a and b, the sugar moiety of codon nucleosides is coordinated past magnesium ion (shown in magenta) together with small subunit elements (Pro 48 of S12 and C518 with G530 in 16S rRNA); coordination distances show that the position of the wobble nucleotide in the codon can slightly arrange depending on the type of pairing interactions. The anticodon nucleotide is weakly restricted by C1054 in 16S rRNA via lone pair–aromatic interactions. The lower panel displays the standard wobble G34·U(+six) pair with specified parameters as in a and b; encounter PDB lawmaking: 3H8I. (c) Superposition of S34·A(+6) and S34·G(+6) pairs by the mRNA codons (left) or by the tRNALys SUU anticodon (right) shows relative extent of the guanosine and modified uracil displacement in the S34·G(+6) pair; the deportation is estimated by angstroms and marked by red arrows. In a and b 2F o−F c electron density maps are contoured at 1.5σ.
The model of tRNALys SUU jump to its second cognate codon AAG demonstrated an unprecedented and hit base-pairing geometry (Fig. 3b). The new S34·K(+6) pair is characterized by the larger C1′–C1′ distance of 11.v Å that exceeds a corresponding distance in a standard Watson–Crick pair by one Å. Withal, interatomic distances between Watson–Crick edges within S34·G(+6) imply the existence of two hydrogen bonds between carbonyl oxygen and N3 atom of modified uracil, and N3 atom and amino grouping of guanosine, respectively (Fig. 3b).
1 of the hypotheses that could explain observed pairing interactions suggested that nether physiological conditions a significant fraction of mnmfivesouthtwoU is present in a zwitterionic class (Fig. 4a)41. This is due to the increased acidity of the N3 proton by the anterior upshot of the protonated methylaminomethyl group, every bit was predicted based on the theoretical estimate of the pYard a (ref. 41). In its neutral form, the modified uracil forms 2 hydrogen bonds same equally in the case for a standard Watson–Crick U·A pair and as we observed for the mnm5due south2U34·A(+6) pair (Fig. 4b). In its deprotonated form, the N3 atom of the uracil becomes a proton acceptor and can grade a hydrogen bond with the amino grouping of guanosine, while the uracil carboxyl group is engaged in another hydrogen bond with the guanosine N3 cantlet (Fig. 4c). Although less probable, the observed unusual design of hydrogen bonds between the modified uridine and guanosine can likewise be rationalized by existence of either rare tautomeric states (Fig. 4d,east) or an alternative zwitterionic land (Fig. 4f) of the modified uridine.
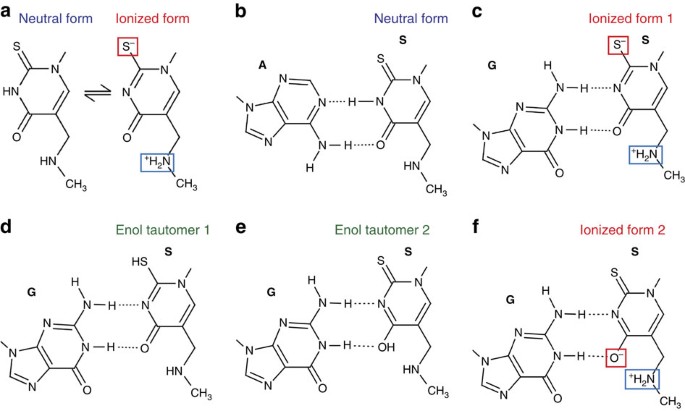
(a) Equilibrium between ii forms of mnm5southward2U41. (b) Interactions with adenosine of the neutral grade of mnmfivesiiU. (c) Theoretically predicted base of operations pair with guanosine of zwitterionic form of mnm5southwardtwoU carrying negative charge on sulfur cantlet. (d,due east) 2 alternative pairing interactions with guanosine and enol tautomeric forms of mnmvdue south2U. (f) Base-pairing interactions with guanosine of a possible zwitterionic form of mnmfivedue southiiU.
It is worth to underline hither that in contrast to the outset 2 codon–anticodon positions, which are tightly restricted by the decoding centre29,30, the 'wobble' pair is not very firmly stabilized. The codon nucleotide is held in place only by indirect interactions with G530, C518 and S12 through a Mg2+ ion (Fig. 3a,b) and the O4' of the starting time anticodon nucleotide forms weak off-centre lone pair–π interaction with the nucleobase of C1054 in 16S rRNA (Supplementary Fig. two)42. Notwithstanding, the fact that observed S34·G(+6) pair is distorted from the standard 'wobble' geometry suggests that the same indirect restraints and interactions of the modification groups provide some restraints to control geometry of the third base pair.
Pyrimidine–pyrimidine mismatch in the 70S decoding centre
The translation of genes into proteins is an fault-decumbent procedure with the average frequencies of mistranslation ten−iii–10−5 (ref. 43). Nosotros have recently published kickoff structural rationales for the phenomenon of translational infidelityxxx. We demonstrated that considering the G·U mismatches can mimic the form of a canonical Watson–Crick pair via tautomerization or ionization, these type of mismatches become accepted in the decoding eye, which restricts geometries of allowed pairs to canonical interactions. At the same time, our models with the A·A and C·A mismatches at the first two positions of the codon–anticodon duplex suggested that these pairs would be efficiently discriminated confronting because of (i) steric clashes inside a mispair or of a mispair with the tight decoding centre itself, or (ii) because of absence of stable pairing interactions between pairing nucleotides29,30.
In the current study, nosotros asked an ensuing question of what is the structural basis for bigotry against the pyrimidine–pyrimidine U·U pair, which represents a low-probability mistake during translation44,45,46. Thus, we determined structures of complexes iii and four (Fig. 1c) where the SUU anticodon of tRNALys SUU formed a U·U mismatch either with the first or the second codon position. As was anticipated, the nucleotides, critical for decoding A1493 and A1492/G530 of 16S rRNA47, stabilized the U·U mismatch at the first and the 2nd codon–anticodon positions via A-pocket-size groove interactions (Fig. 5a). These results substantiated our expanded mechanism of decoding that provides structural basis for discrimination in favour of right tRNAs and against incorrect tRNAs, and describes identical rearrangements of the decoding centre on binding of cognate or virtually-cognate tRNA29,30. In both complexes, the interatomic distances between the Watson–Crick edges of opposing uracils exceeded 3.four Å, implying weak electrostatic interactions (Fig. 5b,c). A hypothetical U·U pair possible through a shift in the keto-enol equilibrium would crave a typical distance of three.0–three.1 Å between uracils for hydrogen bonds to occur (Fig. 5d). Notwithstanding, positions of uracils in both models brand this scenario unlikely (Fig. 5b,c). To put it simply, the restraints on the sugar-phosphate backbones of the codon–anticodon helix imposed by the decoding eye are stiff enough to prevent the uracils to come close plenty to interact strongly via hydrogen bonds (Supplementary Fig. 3).
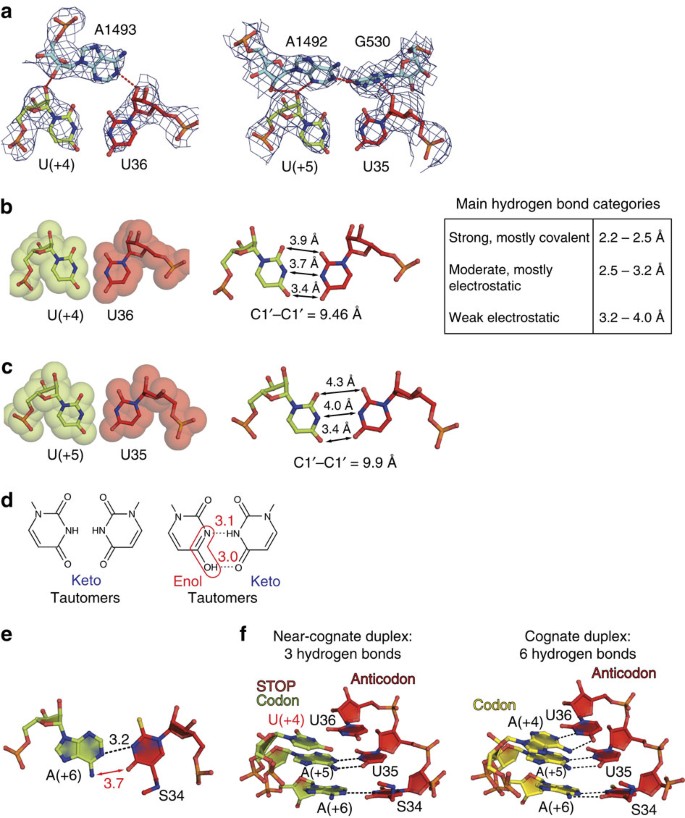
(a) Critical A1492 and A1492/G530 of 16S rRNA stabilize the saccharide phosphate backbones of the U·U mismatch at the first (left) and second (right) positions of the codon–anticodon duplex by A-small groove interactions as is the case for any canonical Watson–Crick pair29,30. 2F o−F c electron density maps are contoured at 1.5σ. (b,c) Geometries of the U·U mismatch at the first (b) and second (c) positions of the codon anticodon duplex; van der Waals surfaces (left) together with interatomic distances (right) are presented. In b, the table describes standard categories of hydrogen bonds. (d) Formation of a strong U·U pair would necessitate a shift in the keto-enol equilibrium from abundant keto form (left console) to a rare enol form (red frame) and the 3-Å distance between Watson–Crick surfaces of opposing uridines (correct panel). (e) Conformation of the wobble S34·A(+6) pair in the complex 3 (see Fig. 1c). The absence of two hydrogen bonds expected for the mnm5s2U·A pair can reflect (i) a deformation of the codon–anticodon minihelix induced by the mismatch and (two) the specific influence of the tRNA modification at position 34 known to annul misreading of the genetic code. (f) The almost-cognate duplex composed of the tRNALys SUU anticodon and the ochre stop codon is significantly weakened compared with the cognate version on the AAA codon with the full set of canonical Watson–Crick interactions. The described weakening of the nearly-cognate duplex would imply dissociation of tRNALys SUU from the ribosome or, in other terms, rejection.
In circuitous 3 with the stop codon UAA and the first U·U mismatch, the codon–anticodon duplex was additionally weakened at the 3′-finish where the S34·A(+vi) pair was slightly deformed (Fig. 5e). As a consequence, only three potent hydrogen bonds were formed betwixt codon and the anticodon compared with the cognate version with six hydrogen bonds (Fig. 5f). These results demonstrated why tRNALys SUU normally does not read the ochre codon. 2 boosted structures of the near-cognate complexes 3 and four (Fig. 1c) solved in the presence of antibiotic paromomycin, which stimulates miscoding, supported our previous conclusion of the antibiotic mechanism of action48. In both cases, paromomycin stabilized A1492 and A1493 in the 'out' from the interior of helix 44 positions, hence stimulating A-small groove interactions with the outset two nucleotides of the A-codon. In addition, binding of the antibiotic led to a positional shift of the A1493 phosphate, resulting in fractional consolation of the restrictive decoding centre from the side of the mRNA codon. Finally, in the ribosome structures with paromomycin we observed a displacement of helix 69 of the large subunit towards the D-stem of tRNALys SUU that, most probably, enhanced stabilization of a nearly-cognate substrate on the ribosome.
It is important to mention that uracils in a mismatch at the first codon–anticodon position were closer to each other than at the 2d position (Fig. 5b,c). In the light of kinetic description of the tRNA option process on the ribosome, information technology tin exist interpreted as the 2d codon–anticodon position being more controlled than the showtime i43. Accordingly, kinetic evaluations of codon readings by tRNALys UUU in bacteria assigned the highest accuracy values for the 2d position in the codon–anticodon duplex45. Thus, information technology is important to indicate here that despite of the fact that we could crystallize described near-cognate states of the ribosome at the proofreading step, in solution these complexes would be prone for dissociation considering of a prominent lack of pairing codon–anticodon interactions.
Discussion
Fifty years agone, Francis Crick suggested some rules for translation of the genetic lawmaking on the ribosome and postulated the wobble hypothesis that gave first explanations to degeneracy of the code8,xviii. Information technology was predicted that the first 2 positions of the codon would pair with the anticodon using the standard base pairs, while in the base pairing of the third codon base 'there is a certain amount of play, or wobble, such that more than one position of pairing is possible'. Get-go examples of foretold wobble not-standard pairs included G·U, U·G, I·A, I·C and I·U pairs, where the nucleoside on the left designates position 34 in tRNAviii. Since those times, a titanic work on deciphering the genetic code resulted into a simple textbook chart where nigh of the codons are two- or fourfold degenerate, pregnant that one amino acid can be coded by two or four codons differing by the third base of operations. Farther identification of tRNA modifications and peculiarly those at the kickoff anticodon 'wobble' position 34 elaborated more than on the phenomenon of degeneracy. The 'modified wobble hypothesis' suggested that specific tRNA base modifications evolved to discriminate particular codons—expanding and facilitating an ability of tRNA to read more than i codon in some cases and preventing misreading in other casesiv.
In the present study, we describe a new type of a base pair at the third wobble position of a codon–anticodon duplex in the 70S ribosome decoding centre. Our structures demonstrate that the reversed 'wobble' pair S34·G(+6) adopts its own geometry, different from the standard G34·U(+6) paireight at the 3rd codon–anticodon position (Fig. 3b). This is maybe due to both certain restraints put on the 3rd base pair by the decoding centre of the 70S ribosome and modifications on the nucleotide S34 of tRNALys SUU, shaping the codon–anticodon helix and the ASL of the tRNA (see Results). In spite of the fact that S34·Chiliad(+half dozen) is not isosteric to the standard wobble pair, at that place is a certain similarity of the overall shape between these two pairs.
Both pairs are characterized past displacement of the anticodon nucleotide—S34 in the S34·Thou(+6) pair and G in the G34·U(+vi) pair—towards the minor groove of the codon–anticodon minihelix. This is achievable because of the upmost location of the nucleotide in the U-turn construction of the tRNA anticodon loop49. On the codon side of the pairs there is another trend of shifting; however, in this case it is towards the major groove of the minihelix. Results reported in this study particularly show this displacement (Fig. 3c right). Hence, a term 'wobble' can be also applied to the chapters of the 3rd codon nucleotide to conform its position because of difference in force and type of restraints imposed by the decoding centre on the 3rd base pair as compared with the start and second base pairs (Fig. 3a,b).
Uridine at the wobble positions of various tRNAs is nigh always modified in bacteria and eukaryotes1,5. In many cases, tRNAs with the modified uridines read two codons catastrophe with purine A or Yard and, in some rare cases, modifications help to recognize all four nucleotides A, Thou, C and U at the third codon position50. Previously, it was shown that preferential form of the third wobble pair with a fully modified uridine approached a standard Watson–Crick-like geometry if this uridine was paired with guanosine26,27 (Supplementary Fig. iv). Thus, in the partial and heterologous model of the isolated bacterial 30S ribosomal subunit, whose crystals were soaked with an ASL of homo tRNA3Lys UUU, the modified mcmfivedue south2U34 formed C·G-like mcm5southii U34·Thousand pair26. In this pair, the uridine base of operations was shifted towards the major groove when compared with geometry of the mnm5s2U·G(+6) pair in our model (Supplementary Fig. 4b left). In another study, uridine-5-oxyacetic acrid at the position 34 of tRNAVal displayed a similar Watson–Crick-similar pair27 with guanosine greatly displaced towards the small groove of a codon–anticodon helix when compared with our structure (Supplementary Fig. 4c left). On the ground of these 30S models, it was reasoned that modifications stabilized enol tautomers of uracil to keep the Watson–Crick-like geometry. It should likewise be mentioned here that although these structures gave important insights into how tRNA modifications can influence base-pairing interactions in the decoding centre, they described a fractional organisation of decoding where roles of a full-length tRNA and the large ribosomal subunit could not be taken into consideration. At the same time, proper agreement of virtually oftentimes subtle fine-tuning effects of tRNA modifications on the codon–anticodon pairing geometries would require advanced experimental organization consisting of total-length ligands and complete ribosome.
The novel pairing interaction at the third position of the codon–anticodon duplex described in this work deepens our understanding of principles embedded into translation of the genetic lawmaking on the ribosome. In agreement with the 'modified wobble hypothesis', our data show that the shape of the 'wobble' base pair is jointly divers past the ribosome environment and the tRNA modifications. In the observed case, such synergy gave ascent to the novel base-pairing design, never observed before in the tRNA–mRNA duplexes. From the observation we can derive both a wider spatial tolerance of the wobble base-pair surround than expected before and, on the other mitt, a certain degree of strictness imposed on the base pair, forcing information technology into the conformation unusual for a relaxed duplex.
Together with our preceding models describing Grand·U, U·G, A·C and A·A mismatches in codon–anticodon duplexes bound in the 70S decoding eye, the present models with pyrimidine–pyrimidine U·U mismatches consolidate the translation fidelity machinery put forward by us earlier29,xxx. In this mechanism, the ribosome responds identically on binding of cognate or near-cognate tRNA by enveloping the codon–anticodon duplex in a rigid universal mould of the 'closed' decoding middle, which favours the Watson–Crick geometry of codon–anticodon base pairs. It is crucial to emphasize that when near-cognate tRNA with a mismatch to the mRNA codon binds to the decoding centre (during initial selection and proofreading steps), the number of hydrogen bonds between the small groove of the nigh-cognate codon–anticodon helix and the 'airtight' decoding center elements is the same as would exist the case during binding of the cognate tRNA. Therefore, the ribosome is incapable to distinguish between these states by the small-scale groove geometry of codon–anticodon base of operations pairs. Still, in contrast to cognate tRNAs that will stably pair to the mRNA codon by canonical Watson–Crick interactions, near-cognate tRNAs with a mismatch to the codon will exist more than likely to dissociate from the ribosome because of the strict restraints imposed by the tertiary construction of tRNA and elements of the 'airtight' decoding centre on the codon–anticodon helix. This pressure constitutes the discriminatory force past preventing the following conformational changes: (a) widening of the codon–anticodon helix needed to adapt bulky non-canonical pairs (for example, A·A pair); (b) narrowing of the codon–anticodon helix necessary for proper pairing interaction in pyrimidine–pyrimidine pairs (for example, U·U pair); or (c) shift of nucleobases towards minor or major groove, characteristic of, for example, wobble M·U pair. The third base pair of the codon–anticodon duplex contributes in a dissimilar manner, compared with the get-go two positions. Its function in decoding is linked to the base pair nature, the indirect restraints imposed by the decoding centre and the presence of the modification groups that influence conformations of the tRNA ASL and the codon–anticodon helix. An additional discriminatory role at the proofreading step can be performed past the tails of some ribosomal proteins that selectively stabilize cognate tRNA substrates51. The rare translational mistakes caused by the incorporation of near-cognate tRNAs are reasoned mostly by the power of some tRNAs to course Watson–Crick-like base pairs via ionization or tautomerism29, or in some cases a mismatch randomly escapes bigotry past preserving geometry shut to the Watson–Crick pairxxx. The present decoding mechanism farther establishes that discrimination betwixt tRNAs is primarily founded on spatial fit29,30,48 rather than on the number of hydrogen bonds between the 'airtight' decoding centre and the codon–anticodon duplex35.
Methods
Materials
Uncharged, native individual tRNALys SUU and tRNAfMet CAU from Eastward. coli were purchased from Chemical Block (Russia). The mRNA constructs whose sequences are specified below were from Thermo Scientific (The states) and deprotected following the supplier procedure. All mRNA constructs contained identical sequence 5′-GGCAAGGAGGUAAAA-3′ at the 5′-end, which was followed by 5′-AUGAAAA6-3′ (mRNA-1), 5′-AUGAAGA9-3′ (mRNA-ii), 5′-AUGUAAAnine-3′ (mRNA-iii) or five′-AUGAUAAix-3′ (mRNA-iv). Aminoglycoside antibiotic paromomycin was purchased from Sigma-Aldrich.
Purification of the ribosomes
Purification of the 70S ribosomes from strain HB8 of T. thermophilus was performed according to the protocol described in ref. 30.
Complex germination
All ribosomal complexes were formed in 10 mM Tris-acetate pH vii.0, 40 mM KCl, 7.five mM Mg(CH3COO)ii, 0.five mM dithiothreitol at 37 °C. For the cognate complexes (Fig. 1c, complexes 1 and 2), the 70S ribosomes (iii μM) were pre-incubated with fivefold backlog of mRNA-1 or mRNA-ii and threefold excess of tRNAfMet CAU for fifteen min to fill up the P-site. Then, tRNALys SUU was added at fivefold backlog and incubation was continued for thirty min. Nigh-cognate complexes (Fig. 1c, complexes iii and four) were prepared in a similar manner with the use of mRNA-3 and mRNA-4 constructs. Complexes with paromomycin were obtained by including the antibiotic (60 μM) into the incubation mixture containing 70S/tRNAfMet/mRNA-iii/tRNALys SUU or 70S/tRNAfMet/mRNA-4/tRNALys SUU.
Crystallization and crystal handling
Crystals were grown at 24 °C via vapour diffusion in sitting-drib plates (CrysChem, Hampton Inquiry). The ribosomal complex (two μl) containing 2.eight mM Deoxy Big Chaps (CalBioChem) was mixed with the equal volume of the crystallization solution (3.9–4.ii% (west/v) PEG 20k, three.9–iv.ii% (w/v) PEG550mme, 100 mM Tris-acetate pH 7.0, 100 mM KSCN). The crystals grew for 2–3 weeks and were so dehydrated past exchanging the reservoir for lx% (v/5) ii-methyl-ii,4-pentanediol. Before freezing in the nitrogen stream, crystals were and then cryoprotected by the addition of xxx% (v/5) two-methyl-ii,four-pentanediol and xiv mM Mg(CHthreeCOO)2.
Construction determination
Information for all complexes were collected at the PXI beamline of Swiss Calorie-free Source, Switzerland, at 100 K. A very depression-dose manner was used and high redundancy information were collected52. The information were processed and scaled using XDS53. All crystals belong to space group P21two12one and contain two ribosomes per asymmetric unit of measurement. One of the previously published structures29, with tRNA, mRNA and metallic ions removed, was used for refinement with Phenix54. The initial model was placed within each data set by rigid body refinement with each biopolymer chain as a rigid body. This was followed by initial coordinate refinement. The resulting electron density maps were inspected in Coot55, and the tRNA and mRNA ligands were built in. During several cycles of manual rebuilding followed past coordinate and isotropic B-factor refinement, magnesium ions were added and the last refinement round took place. The data collection and refinement statistics are presented in Table i.
Additional information
Accretion codes: The atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Information Banking company under accession codes: 5E7K, 5E81, 5EL4, 5EL5, 5EL6 and 5EL7.
How to cite this article: Rozov, A. et al. Novel base of operations-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 7:10457 doi: 10.1038/ncomms10457 (2016).
Accession codes
Accessions
Poly peptide Data Bank
References
-
Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
-
El Yacoubi, B., Bailly, G. & de Crecy-Lagard, 5. Biosynthesis and role of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 46, 69–95 (2012).
-
Nishimura, S. & Watanabe, K. The discovery of modified nucleosides from the early days to the nowadays: a personal perspective. J. Biosci. 31, 465–475 (2006).
-
Agris, P. F., Vendeix, F. A. & Graham, Due west. D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366, 1–13 (2007).
-
Cantara, W. A. et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 39, D195–D201 (2011).
-
Durant, P. C., Bajji, A. C., Sundaram, M., Kumar, R. K. & Davis, D. R. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the outcome of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44, 8078–8089 (2005).
-
Stuart, J. W., Koshlap, K. M., Guenther, R. & Agris, P. F. Naturally-occurring modification restricts the anticodon domain conformational space of tRNA(Phe). J. Mol. Biol. 334, 901–918 (2003).
-
Crick, F. H. Codon—anticodon pairing: the wobble hypothesis. J. Mol. Biol. nineteen, 548–555 (1966).
-
Yarian, C. et al. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 277, 16391–16395 (2002).
-
Bjork, Thou. R. et al. Transfer RNA modification. Annu. Rev. Biochem. 56, 263–287 (1987).
-
Agris, P. F. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie 73, 1345–1349 (1991).
-
Beuning, P. J. & Musier-Forsyth, K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers 52, ane–28 (1999).
-
Grosjean, H. & Bjork, Grand. R. Enzymatic conversion of cytidine to lysidine in anticodon of bacterial isoleucyl-tRNA--an alternative way of RNA editing. Trends Biochem. Sci. 29, 165–168 (2004).
-
Bjork, 1000. R. et al. Transfer RNA modification: influence on translational frameshifting and metabolism. FEBS Lett. 452, 47–51 (1999).
-
Urbonavicius, J., Qian, Q., Durand, J. One thousand., Hagervall, T. Chiliad. & Bjork, G. R. Comeback of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873 (2001).
-
Thiaville, P. C. & de Crecy-Lagard, V. The emerging role of complex modifications of tRNA in signaling pathways. Microb. Cell 2, 1–4 (2015).
-
Kellogg, D. A., Doc, B. P., Loebel, J. Eastward. & Nirenberg, Thou. Westward. RNA codons and protein synthesis. Ix. Synonym codon recognition by multiple species of valine-, alanine-, and methionine-sRNA. Proc. Natl Acad. Sci. USA 55, 912–919 (1966).
-
Crick, F. H. The origin of the genetic code. J. Mol. Biol. 38, 367–379 (1968).
-
Raba, M. et al. Nucleotide sequence of 3 isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur. J. Biochem. 97, 305–318 (1979).
-
Sprinzl, 1000. & Vassilenko, K. S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33, D139–D140 (2005).
-
Armengod, M. Eastward. et al. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie 94, 1510–1520 (2012).
-
Kumar, R. Grand. & Davis, D. R. Synthesis and studies on the effect of 2-thiouridine and four-thiouridine on carbohydrate conformation and RNA duplex stability. Nucleic Acids Res. 25, 1272–1280 (1997).
-
Ogle, J. M. et al. Recognition of cognate transfer RNA past the 30S ribosomal subunit. Scientific discipline 292, 897–902 (2001).
-
White potato, F. 5. T. & Ramakrishnan, V. Structure of a purine-purine wobble base pair in the decoding heart of the ribosome. Nat. Struct. Mol. Biol. xi, 1251–1252 (2004).
-
Murphy, F. Five. t., Ramakrishnan, V., Malkiewicz, A. & Agris, P. F. The function of modifications in codon bigotry by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. xi, 1186–1191 (2004).
-
Vendeix, F. A. et al. Human tRNA(Lys3)(UUU) is pre-structured past natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J. Mol. Biol. 416, 467–485 (2012).
-
Weixlbaumer, A. et al. Machinery for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 14, 498–502 (2007).
-
Jenner, L. B., Demeshkina, Due north., Yusupova, Grand. & Yusupov, M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560 (2010).
-
Demeshkina, N., Jenner, L., Westhof, East., Yusupov, M. & Yusupova, One thousand. A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012).
-
Rozov, A., Demeshkina, N., Westhof, E., Yusupov, Grand. & Yusupova, K. Structural insights into the translational infidelity mechanism. Nat. Commun. 6, 7251 (2015).
-
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
-
Fernandez, I. S. et al. Unusual base of operations pairing during the decoding of a terminate codon by the ribosome. Nature 500, 107–110 (2013).
-
Schmeing, T. Yard., Voorhees, R. Yard., Kelley, A. C. & Ramakrishnan, V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol. xviii, 432–436 (2011).
-
Holley, R. Westward. et al. Structure of a ribonucleic acid. Science 147, 1462–1465 (1965).
-
Ogle, J. G., Murphy, F. 5., Tarry, M. J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
-
Yokoyama, South. et al. Molecular machinery of codon recognition by tRNA species with modified uridine in the beginning position of the anticodon. Proc. Natl Acad. Sci. USA 82, 4905–4909 (1985).
-
Mazumdar, Southward. K. & Saenger, W. Molecular structure of poly-ii-thiouridylic acid, a double helix with not-equivalent polynucleotide chains. J. Mol. Biol. 85, 213–219 (1974).
-
Demeshkina, N., Jenner, L., Yusupova, M. & Yusupov, M. Interactions of the ribosome with mRNA and tRNA. Curr. Opin. Struct. Biol. xx, 325–332 (2010).
-
Nakamura, Y. Codon Usage Database http://world wide web.kazusa.or.jp/codon/ (2007).
-
Frédéric Dardel, F. K. Bioinformatics: Genomics and Postal service-Genomics John Wiley & Sons, Ltd. (2006).
-
Takai, K. & Yokoyama, S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 31, 6383–6391 (2003).
-
Egli, M. & Sarkhel, Southward. Lone pair-aromatic interactions: to stabilize or not to stabilize. Acc. Chem. Res. 40, 197–205 (2007).
-
Wohlgemuth, I., Pohl, C., Mittelstaet, J., Konevega, A. L. & Rodnina, M. 5. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2979–2986 (2011).
-
Kramer, E. B. & Farabaugh, P. J. The frequency of translational misreading errors in East. coli is largely determined past tRNA contest. RNA thirteen, 87–96 (2007).
-
Johansson, Thousand., Zhang, J. & Ehrenberg, Yard. Genetic lawmaking translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc. Natl Acad. Sci. United states 109, 131–136 (2012).
-
Manickam, N., Nag, N., Abbasi, A., Patel, Yard. & Farabaugh, P. J. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against virtually potential errors. RNA 20, 9–15 (2014).
-
Moazed, D. & Noller, H. F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J. Mol. Biol. 211, 135–145 (1990).
-
Demeshkina, N., Jenner, 50., Westhof, East., Yusupov, M. & Yusupova, One thousand. New structural insights into the decoding mechanism: translation infidelity via a M.U pair with Watson-Crick geometry. FEBS Lett. 587, 1848–1857 (2013).
-
Westhof, E., Yusupov, Chiliad. & Yusupova, One thousand. Recognition of Watson-Crick base pairs: constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. half-dozen, 19 (2014).
-
Grosjean, H., de Crecy-Lagard, Five. & Marck, C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 584, 252–264 (2010).
-
Jenner, 50., Demeshkina, North., Yusupova, One thousand. & Yusupov, Yard. Structural rearrangements of the ribosome at the tRNA proofreading footstep. Nat. Struct. Mol. Biol. 17, 1072–1078 (2010).
-
Mueller, M., Wang, M. & Schulze-Briese, C. Optimal fine phi-slicing for single-photon-counting pixel detectors. Acta Crystallogr. D Biol. Crystallogr. 68, 42–56 (2012).
-
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
-
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
-
Emsley, P., Lohkamp, B., Scott, Westward. Thou. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
-
Karplus, P. A. & Diederichs, K. Linking crystallographic model and information quality. Science 336, 1030–1033 (2012).
Acknowledgements
We thank the staff of the PXI beamline at the Swiss Light Source (Switzerland) for the advices in the synchrotron data collection. We are also thankful to Henry Grosjean for disquisitional reading of the manuscript and constructive discussions. This work was supported past the French National Research Agency ANR-11-BSV8-006 01 (to K.Y.), the European Research Council advanced grant 294312 (to M.Y.), the Foundation for Medical Enquiry in France grant FDT20140930867 (to I.K.) and the Russian Regime Program of Competitive Growth of Kazan Federal University (to I.One thousand., M.Y. and K.Y.). This work was also supported past the LABEX: ANR-x-LABX-0036_NETRNA and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the time to come program (to E.Due west.).
Author data
Affiliations
Contributions
A.R. and Due north.D. conducted the experiments. I.One thousand. purified ribosomes and assisted in X-ray data collection. A.R., Northward.D., E.W. and G.Y. interpreted the structures. A.R., North.D., I. K., East.Due west., M. Y. and Yard.Y. contributed to the last version of the paper.
Respective author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed nether a Creative Commons Attribution four.0 International License. The images or other 3rd party material in this article are included in the commodity's Creative Commons license, unless indicated otherwise in the credit line; if the fabric is not included nether the Creative Commons license, users will demand to obtain permission from the license holder to reproduce the textile. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Reprints and Permissions
Well-nigh this article
Cite this article
Rozov, A., Demeshkina, North., Khusainov, I. et al. Novel base of operations-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun 7, 10457 (2016). https://doi.org/x.1038/ncomms10457
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/ncomms10457
Further reading
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you lot find something abusive or that does not comply with our terms or guidelines delight flag information technology equally inappropriate.
Using Wobble Rules, What Is the Minimum Number of Trna Needed to Read the Genetic Code
Source: https://www.nature.com/articles/ncomms10457
0 Response to "Using Wobble Rules, What Is the Minimum Number of Trna Needed to Read the Genetic Code"
Post a Comment